Learn about our proprietary cancer simulation tool
used to improve drug development success
What is Virtual Tumour™?
Virtual Tumour™ is our predictive quantitative systems pharmacology (QSP) model used worldwide by global pharmaceuticals to small biotech companies to support oncology R&D.
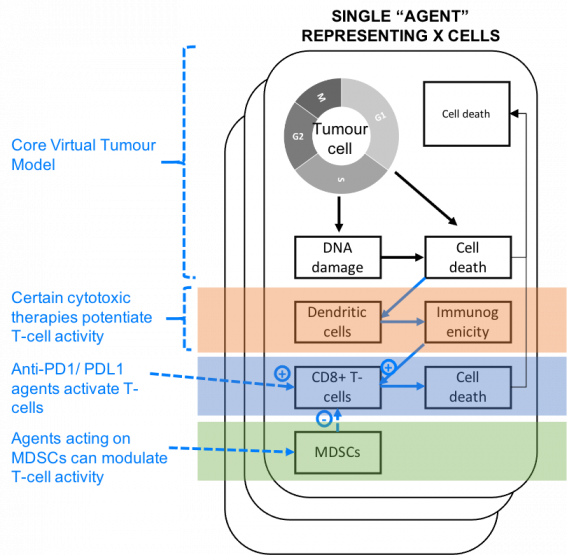
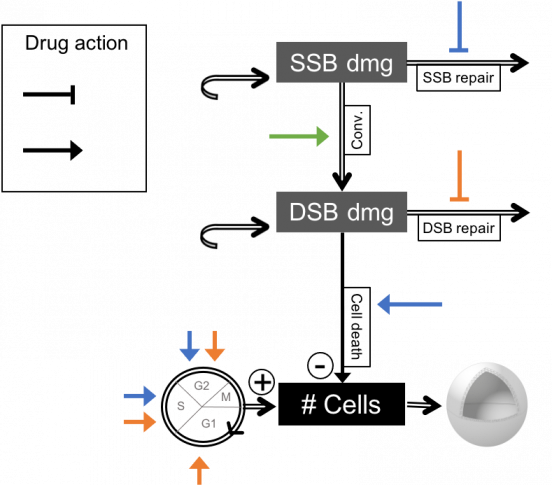
Virtual Tumour is a semi-mechanistic model that incorporates an abstracted representation of the cell cycle, DNA damage/ repair and elements of the immune response.
Virtual Tumour is calibrated using available PK and PD data and can predict the effect on tumour growth of combinations of anti-cancer agents, including radiation.
How Does it Work?
Virtual Tumour™ works by predicting the effects of cancer treatment regimes, simulating the growth or shrinkage of tumours under the effects of any combination of anti-cancer agents.
1
Use established or de novo PK models. Source tumour growth Inhibition/biomarker data from client or public domain. Characterize MOA of agent(s).
2
Model key elements of cell cycle. Calibrate model using available data. Follow cancer cells over time as they die or divide.
3
Track the number of cells and mathematically distribute them in the growing layer of a tumour.
4
Track the tumour size / cell cycle phases / biomarker evolution over time.Predict effect of different regimens.
Virtual Tumour Mechanistic Extensions
Our core Virtual Tumour™ model has been extended to incorporate a variety of additional mechanisms employed in developing cancer treatments.
This project was presented in a poster at AACR.
Contact us for further information on this model.
Why Virtual Tumour™ is a Partner of Choice in Oncology Drug Development?
Our Virtual Tumour™ platform is one of the industry’s most advanced predictive models focused solely on oncology, predicting optimal drug dosing, scheduling and combinations.
Key Features
| Features | Supported |
|---|---|
| Predict behaviour over a full time course, not just a single point in time | |
| Powerfully and naturally combine the effects of drugs with different mechanism of actions (MOAs) | |
| Allows complex dosing and scheduling options e.g. dosing holidays, to be simulated | |
| New data can be quickly incorporated as it is generated in mouse or human trials | |
| Can help explain the behaviour of combination regimes in terms of cellular effects (e.g. cell cycle synchronization, cell death, DNA damage) | |
| Model revisions and additional simulations can be delivered in real-time to enable decision-making during early clinical trials. |
Things You Should Know About Virtual Tumour™
Virtual Tumour™ Explained In Just 90 Seconds
Watch our quick overview of Virtual TumourTM and how our unique system can help you deliver results
Interested in speaking with a consultant?
We’ll arrange a brief consultation for you with a specialist in our team.
SET UP CONSULTATION


